What follows is my version of the mtDNA tree found at PhyloTree. The main characteristic is that each 10 horizontal pixels in the full size image represent one (known) SNP.
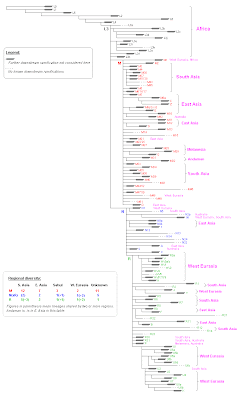
Click to enlarge
Anotations in magenta at the right are merely informative of the main location of the various lineages. I used only 7 distinct geographical regions for this purpose: Africa, West Eurasia (extending sometimes to North Africa and Central Asia), South Asia (extending in one case to Central Asia), East Asia (extending sometimes through North Eurasia, Central Asia and America), Andaman Islands, Australia and Melanesia. Some lineages I really don't know right now where they have been found and have been left without anotations.
Preliminary findings: the rythm of expansion.
Very simplified, I find the following apparent pulses:
1. A very old one when L0, L1 and L2'3'4'5'6 begin their expansion.
2. A "rapid" sequence of expansion of L3:
2.1. Expansion of L3 and L2.
2.2. Expansion of three L3 subclades: L3b'c'd'j, L3e'ik'x and L3h.
2.3. Expansion of M, N and many M sublineages of "very short stem" (1 SNP)
2.4. Expansion of R and many M, N and R sublineages of "short stem". Includes also N1, N9, S, most Melanesian M, most South Asian M that had not expanded before, M7, M9, D...
3. After a brief "pause", expansion of the main West Eurasian lineages: HV and U, coincident with some les relevant branching pulses in South and East Asia and even Melanesia.
4. The latest multiple expansive pulse I have located, apparently some time after step 3, is centered in East Asia and surely related to the colonization of the north. The main clades taking part are C and Z, but also M10, M11, M12 and M13 (all East Asian, except the last that I don't know right now). It has some parallel in West Eurasia with the expansion of many U subclades, notably U5, U6 and R0a.
Update: I had to correct an anotation in the image and used that intervention to add a table of diversity of the three major Eurasian macro-haplogroups by regions. It is very clear that M and R diversity gravitates around South Asia. The case of N(xR) is less clear and should depend in where the various minor N lineages are found maybe. Still the case for a South Asian origin can be made for this clade too, moreso if we add R to the figure of South Asian N clades (all three shared with other regions though).
.









4 comments:
Good one Maju. But I'm pretty sure haplogroup S is found in Australia, not Melanesia. They are different places you realise? I also can't see the old haplogroup I. I presume it's part of the N1 clade.
Good one Maju. But I'm pretty sure haplogroup S is found in Australia, not Melanesia.Oh, man! I was confused (too much data at the same time!): had it tagged as Austrlian and yesterday night I happened to think "damn, no it's Melanesian!" and "corrected" it.
Will restore the previous version. Thanks.
I also can't see the old haplogroup I. I presume it's part of the N1 clade.Sure: I is a subclade of a subclade of N1. It is a small lineage not worth mentioning in such a large tree.
"Oh, man! I was confused (too much data at the same time!)".
What makes it more confusing is that mtDNA S is Australian and YDNA S is, at least mainly, New Guinea. In fact I'm not sure if it's found in Australia at all.
YDNA S is found in Australia,but at very low levels.
Post a Comment